4D Cell-ECM interactions in multicellular systems
Our laboratory studies how the extracellular matrix (ECM) functions as a dynamic information matrix that regulates tissue morphogenesis and regeneration in mammalian skin. Using ECM molecular mapping, quantitative 4D (3D + time) imaging, time-resolved single-cell transcriptomics and a variety of perturbations, we uncover how molecular composition, organisation and mechanics of the ECM shape tissue architecture over time. Our long-term goal is to engineer cell–ECM interactions to control morphogenesis and patterning.

1. ECM spatiotemporal code
Spatiotemporal information encoded in the ECM
We investigate how the spatiotemporal organisation of the ECM encodes information that defines distinct microenvironments for stem cells and their niche cells. Through comprehensive ECM mapping and functional studies, we have shown that specific ECM molecules act as molecular “spatiotemporal codes” that selectively mediate epithelial stem/progenitor–niche interactions, thereby regulating tissue development and regeneration (Fujiwara et al., Cell 2011; Cheng et al., eLife 2018; Tsutsui et al., Nat Commun 2021; Liu et al., J Invest Dermatol 2025). This extracellular atlassing framework underpins multiple projects in our lab and continues to generate new research directions. We are now moving towards integrating ECM spatial mapping with spatial transcriptomics to link ECM codes to cell states across development, regeneration and disease.

2. ECM dynamics
Multi-scale ECM dynamics in 4D
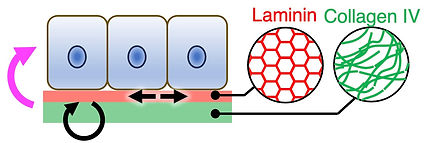
The ECM is no longer viewed as a static scaffold formed by the passive deposition of proteins, but rather as a dynamic polymer network that is critical in morphogenesis and regeneration. However, how ECM dynamics at molecular level are spatiotemporally regulated, and how these dynamics are coupled to tissue-scale growth and mechanics remains poorly understood.
To visualise ECM dynamics in situ, we developed fluorescent basement membrane reporter mouse lines that enable long-term, quantitative 4D visualisation of basement membrane dynamics in living mammalian tissues (Wuergezhen et al., J Cell Biol 2025). Using this platform, we revealed that spatially patterned molecular turnover of collagen IV is coupled to macroscopic basement membrane expansion and anisotropic epithelial tissue growth. In parallel, we are advancing quantitative data-driven mathematical analyses and modelling approaches to describe and predict the relationship between molecular turnover, basement membrane expansion and mechanical properties. We are further expanding our imaging toolkit to characterise the basement membrane as a self-organising, active polymer network, in which patterned molecular organisation and turnover dynamically tune tissue mechanics and cell behaviours. We aim to understand how basement membrane–epithelial interactions enable multicellular systems to generate tissue architectures beyond the capacity of cells alone.

3. Development & regeneration
Emergence of tissue architecture through cell–ECM interactions
Skin is a dynamic interface between organisms and the environment, and its architecture varies widely across species to support adaptation. During development, skin appendages, such as hair follicles and feathers, emerge from a 2D epithelial sheet to form diverse 3D structures and to generate stem cells that enable cyclic regeneration and context-dependent responses. Yet, despite extensive work on individual appendages, the general principles that produce such morphological diversity across development, regeneration and evolution remain unclear.
To address this gap, we combined a quantitative 4D imaging platform with time-course single-cell transcriptomics to capture the developmental dynamics underlying mouse hair follicle morphogenesis (Morita et al., Nature 2021). This approach revealed that hair follicle epithelial architecture emerges through a transition from a 2D concentric pre-pattern to 3D cylindrical functional domains. Importantly, this analysis identified the outer ring zone of the concentric pre-pattern as the developmental origin of hair follicle epithelial stem cells. We define this coordinated, concentric-to-cylindrical developmental trajectory as a telescope-type morphogenetic process, in which collective cell behaviours progressively reorganise tissue architecture. By extending this telescopic developmental framework to other skin appendages through cross-organ and cross-species comparison, we aim to uncover general principles by which coordinated cell behaviours and their interactions with the ECM generate robust yet diverse tissue architectures during development, regeneration and evolution.


4. Technology & tool development
Quantitative 4D mapping, imaging and manipulation across scales
These studies are enabled by technologies and tools we developed to atlas, image, and quantify cells and ECM at single-cell resolution in 4D. The major methods and tools established to date are summarised below. We further aim to go beyond observation and analysis by developing novel artificial proteins and advanced genetic manipulation strategies to design and manipulate cell–ECM interactions.
-
An ECM atlas framework that enables spatial mapping of the molecular composition and structural properties of the basement membrane (Tsutsui et al., Nat Commun 2021)
-
An integrated single-cell-resolution long-term 4D imaging and time-course transcriptomics to reconstruct cell lineage dynamics in developing tissues (Morita et al., Nature 2021)
-
Endogenous fluorescent collagen IV knock-in mouse models for quantitative 4D basement membrane imaging in living tissues (Wuergezhen et al., J Cell Biol 2025)
-
A single-cell trannscriptomics–based approach to reconstruct the temporal axis of cyclic tissue regeneration (e.g., reconstruction of a pseudo-hair cycle) (Yokota et al., Cell Rep 2025)
Our goal
Our long-term goal is to elucidate the operating principles of the cellular microenvironment and, based on this understanding, to design and manipulate the microenvironment in order to control of biological phenomena from outside the cell.
